GETTING CLONED GENES INTO BACTERIA BY TRANSFORMATION
Once the gene of interest is cloned into a vector, the construct can be put back into a bacterial cell through a process called transformation (Fig. 3.18) (see Box 3.2).
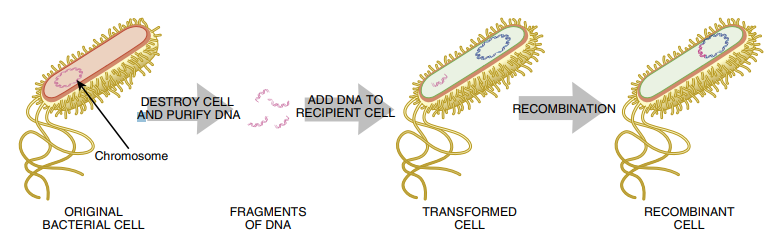
FIGURE 3.18 Transformation Bacterial cells are able to take up DNA such as recombinant plasmids by incubation with metal ions such as Ca++ on ice. This destabilizes the bacterial cell wall, and after a brief heat shock, some of the bacteria take up the DNA or plasmid. If the DNA integrates in the chromosome, the recombinant cell will express any genes found on the DNA. Whole plasmids can also be taken up by a bacterial cell, and these exist as extrachromosomal elements with their own origin of replication.
destabilize 불안정하게 만들다
Here, the DNA construct is mixed with competent E. coli cells. To make the cells competent, that is, able to take up DNA, the cell wall must be temporarily opened up. E. coli cells are mixed with calcium ions on ice and then shocked at a higher temperature such as 42°C for a few minutes, which destabilizes the membrane and cell wall. Most of the cells die during the treatment, but some survive and take up the DNA.
competent 만족할 만한
Another method to make E. coli cells competent is to expose them to a high-voltage shock. Electroporation opens the cell wall, allowing the DNA to enter. This method is much faster and more versatile.
Electroporation is used for other types of bacteria as well as yeast. Bacteria can have different plasmids, but they must have different origin of replications. If there are two different plasmids with the same origin of replication, one of the two will be lost during bacterial replication.
For example, if genes A and B are both cloned into the same kind of vector and both cloned genes get into the same bacterial cell, the bacteria will lose one plasmid and keep the other. This is due to plasmid incompatibility, which prevents one bacterial cell from harboring two of the same type of plasmid.
incompatibility 양립할 수 없음
Incompatibility stems from conflicts between two plasmids with identical or related origins of replication. Only one is allowed to replicate in any given cell. If a researcher wants a cell to have two cloned genes, then two different types of plasmids could be used, or both genes could be put onto the same plasmid.
conflict 갈등
identical 동일한
CONSTRUCTING A LIBRARY OF GENES
Gene libraries are used to find new genes, to sequence entire genomes, and to compare genes from different organisms (Fig. 3.19). Gene libraries are made when the entire DNA from one particular organism is digested into fragments using restriction enzymes, and then each of the fragments is cloned into a vector and transformed into an appropriate host. The basic steps used to construct a library are as follows:
1. Isolate the chromosomal DNA from an organism, such as E. coli, yeast, or humans.
2. Digest the DNA with one or two different restriction enzymes.
3. Linearize a suitable cloning vector with compatible restriction enzyme(s) sites.
4. Mix the cut chromosome fragments with the linearized vector and ligate.
5. Transform this mixture into E. coli.
6. Isolate large numbers of E. coli transformants
Linearize 선으로 만들다
compatible 호환이 되는
ligate 묶다
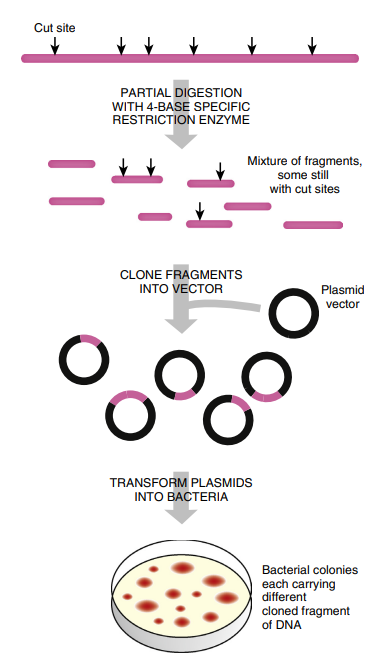
FIGURE 3.19 Creating a DNA Library Genomic DNA from the chosen organism is first partially digested with a restriction enzyme that recognizes a four base-pair sequence. Partial digestions are preferred because some of the restriction enzyme sites are not cut, and larger fragments are generated. If every recognition site were cut by the restriction enzyme, then the genomic DNA would not contain many whole genes. The genomic fragments are cloned into an appropriate vector, and transformed and maintained in bacteria.
The type of restriction enzyme affects the type of library. Because restriction sites are not evenly spaced in the genome, some inserts will be large and others small.
Using a restriction enzyme that recognizes only four base pairs will give a mixture of mostly small fragments, whereas a restriction enzyme that has a six or eight base-pair recognition sequence will generate larger fragments. (Note that finding a particular four base-pair sequence in a genome is more likely than finding a six base-pair sequence.)
Even if an enzyme that recognizes a four base-pair recognition sequence is used to digest the entire genome, these sites are not equally spaced throughout the genome of an organism.
The digested genome will contain some segments too large to be cloned, and some segments too small. To avoid cutting pieces too small, partial digestion is often used. The enzyme is allowed to cut the DNA for only a short time, and many of the restriction enzyme sites are not cut, leaving larger pieces for the library. In addition, it is customary to construct another library using a different restriction enzyme.
Gene libraries are used for many purposes because they contain almost the entire genome of a particular organism.