INTRODUCTION
Numerous plasmids of gram-positive bacteria have been identified, analyzed for their properties, and used as cloning vectors. The range of species from which plasmids have been isolated is broad, yet the properties of most of these plasmids indicate that a common gene pool had been used in their formation (36, 76).
The regions of homology may involve essential or nonessential genes, and a single plasmid has composite character, comprising the antibiotic resistance gene of one plasmid, the replication region of another, etc. The relatedness of 14 of these plasmids is shown schematically in Fig. 1.
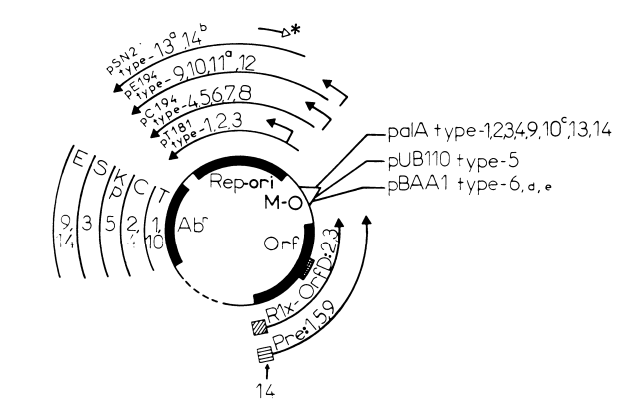
FIG. 1. Regions of relatedness among 14 ssDNA plasmids. A prototype ssDNA plasmid is shown in the inner circle, showing four functions commonly present. Plasmids listed together on the outer lines or arrows have homology in the corr-esponding function, as deduced by DNA and/or amino acid comparison. Plasmids are as follows: 1. pTlX81 2, pC221; 3, pS194: 4, pC194; 5, pUBIIO; 6. pBAA1; 7, pCB101; 8, plIlO1; 9, pE194; 10, pMK158; 11, pADB201: 12. pSH71: 13, pSN2; 14, pIM13 (for further details, see Table 1). Abbreviations and symbols: Rep, replication protein required for plus origin (ori) activity: *-. origin, either embedded or upstream of rep. and direction of replication; -, uncharacterized plus origin for which the dir-ection of replication is deduced by the active orientation of the minus origin pa/A; Abr, antibiotic resistance (T. tetracycline: C. chloramnphenicol: K. kanamycin. P. phleomycin; S. streptomycin; E, erythromycin): Orf, open reading frame(s), encoding either Pre (protein for- recombination 1181) and its target site, RS,, just upstream (M) (18), or Rlx and OrfD, overlapping polypeptides involved in the formation of a relaxation complex at a site just upstream (0) (76); MM, coOp, a locus analyzed only for pTl81 (19, 20, 35) and pUB110 (Polak, personal communiccation). The order of functions on this circular representation is not fixed. ----- on the primary plasmid outline represents unrelated Or uncharacterized sequences which may be present. "pSN2 and pADB201 have been fully sequenced, and each contains only one open reading frame larger than 60 amino acids, presumed or inferred to be involved in replication; ha plasmid identical to pIM13, pNE131. has been isolated from S. epidlertnitli.s (48, 75); cthe M-0 of pMK158, a streptococcal plasmid, has been described as pa/A-like (12), but the pal/A M-O (24) from a staphylococcatl plasmid is nonfunctional in Streptococcus spp. (12); 'I.Cthe M-O of plasmid pBAA1 (14) is also present on Bacillus plasmids pLS11 (10) and pTA1060 (5).
How did such a highly interrelated yet widespread family of replicative elements arise? A recent finding provides a clue; the plasmids under study, of which more than a dozen are already sequenced, all replicate via a single-stranded deoxyribonucleic acid (ssDNA) intermediate, probably by rolling-circle replication (RCR) (82, 83). This particular group of plasmids is referred to here as ssDNA plasmids.
The production of ssDNA and the RCR mechanism have significant consequences for the recombination capacity of these plasmids; both homologous and illegitimate recombination are greatly stimulated, compared with frequencies observed in the chromosome. The high recombination capacity of these plasmids may accelerate their dissemination and explain why they are so interrelated.
Discrete classes of recombination events are shown to occur as a direct consequence of RCR, i.e., homologous recombination between long homologous repeats (61); illegitimate recombination between short direct repeats (7, 38, 69; L. Janniere and S. Ehrlich, submitted for publication); formation of linear multimeric species of plasmids carrying certain DNA insertions in a wild-type background (23); recombination arising from aberrant replicative initiation or premature termination (22, 53); and, in some plasmids, a site-specific recombination system for which the effects of plasmid replication were not analyzed (18). The ssDNA plasmids represent an important family of replicons.
The purpose of this article is to simplify future plasmid analyses by describing their characteristics, with emphasis on the unifying features. Methods of identifying new members of this family are given. Classification of plasmids according to their mode of replication should prove useful in the construction of cloning vectors. It should be noted that little is known of plasmids that are isolated from gram-positive bacteria and that are not of the ssDNA type. However, certain plasmids which do not share any of the common features identified for ssDNA plasmids probably replicate differently.
For those that have been tested, e.g., plasmids pAMP1 and pTB53 (Janniere and Ehrlich, submitted), differences in behavior were noted. Several recent reviews may be of interest, since they emphasize different characteristics of the ssDNA replicons. References 2 and 58 discuss the replication of ssDNA bacteriophages; references 63, 66, and 76 and R. Novick, Annu. Rev. Microbiol., in press, discuss the organization and function of ssDNA plasmids of Staphvlococcas alelacs; and reference 17 discusses recombination in ssDNA plasmids.
PLASMID REPLICATION
Replication Mechanism for the Normal Case
For two plasmids, pTlI81 and pC194, detailed studies have demonstrated thcat replication occurs by RCR (22. 40, 45).
The high degree of homology of the Rep proteins and plus origins of these two plasmids with those of at least seven other sequenced plasmids strongly suggests that all these plasmids replicate in a similar way, i.e., by RCR (22, 76). The RCR mechanism has been extensively studied for ssDNA Escheriuchia (oli bacteriophages (see reference 2 for a review on mechanism).
Three plasmid-encoded elements are used for RCR: a plus origin, a replication protein (Rep), and a minus origin (M-O). The replication mechanism is shown schematically in Fig. 2.
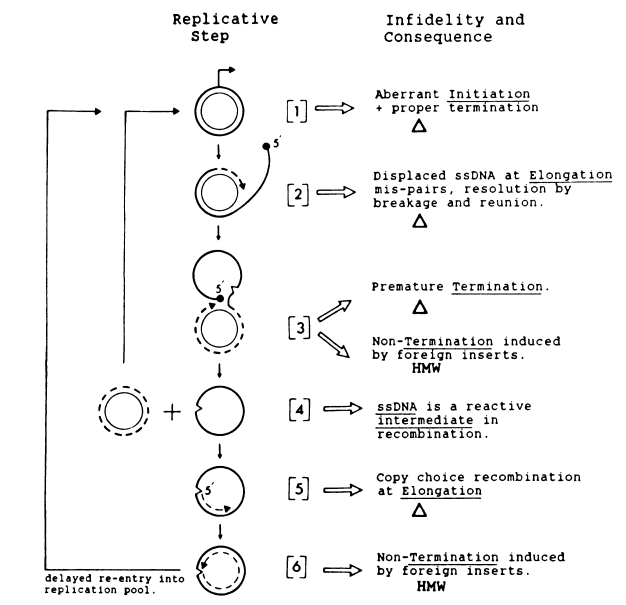

Rep (shown for pT181 to have topoisomerase [i.e., nickingclosing] activity [451) recognizes an origin sequence and produces a nick to initiate replication (step 1). lThis event triggers displacement of the plus strand and polymerization of a new plus strand by 3'-OH extension from the nick (step 2). Then Rep recognizes a termination sequence, which overlaps the origin sequence, and produces a second nick to generate one fully replicated strand and an ssDNA monomer of the displaced strand (step 3).
Finally, Rep ligates the ends of the ssDNA to form a circle, which is detectable as a free molecule (step 4). Ihe nicks that initiate and terminate a round of plus-strand synthesis occur at the same site (2, 22). The M-O serves as an efficient initiation site, recognized by host factors, for the conversion of circular plus-strand ssDNA to double-stranded DNA (dsDNA) (step 5). The formation of the dsDNA plasmid product (step 6) marks the completion of one productive cycle of replication, in which two placsmids are generated from a single one.
If the plus strand is not converted to dsDNA, replication will be nonproductive and will result in the synthesis of one dsDNA plasmid and the accumulation of ssDNA.
Plus origins.
Numerous plus origins have been analyzed. They are localized either upstream of, or within, the Rep open reading frame (Fig. 1). The fully active plus origins of pC194 and pTl81 have been localized to 55 and 70 base pairs (bp), respectively (17a, 22).
As mentioned above, the plus origin is recognized twice by Rep in the replication cycle, first for initiation and again, after the plus strand is displaced and a new strand is synthesized, for termination.
The initiation and termination recognition sites of analyzed origins are overlapping but nonidentical, as demonstrated for the analogous class of E. coli ssDNA phages (2), as well as for ssDNA plasmid pC194 (22).
Most origins, with the exception of the pUBIIO origin, contain sequences having a potential for secondary hairpin structures. It is not known whether such structures play a role in replication and, if so, whether they are recognized at the initiation or termination step. The plus origins thus far identified are grouped into three types, according to homologies with other plus origins.
The strongest homologies within the plus origins (Fig. 3) appear in the regions surrounding the nick sites (indicated in Fig. 3). Consensus sequences were also found between E. coli ssDNA phage origins and certain ssDNA plasmid origins (22).
All pairs of plasmids having homologous plus origins also have corresponding homologies (sometimes less stringent) in their Rep proteins. Furthermore, the amino acid motif around the active site of the 4XX174 Rep protein (84) is conserved in the Rep proteins of S. aiuirelis plasmids pC194, pUBIIO (22), and other analyzed plasmids sharing the consensus origin sequence (Fig. 4), suggesting that the mechanism of nicking at the active site between these plasmids and 4)X174 is analogous.
A search of the published sequence (39) of Strepto;nyces lividans ssDNA plasmid pIJiOl (39, 71; J. Pigac, V. Gamulin, D. Vajaklija, Z. Toman, and H. Schrempf, Abstr. 5th Int. Symp. Genet. Ind. Microorg., p. 41, 1986) revealed a similar amino acid motif in the Rep protein (Fig. 4). The Streptomyces ssDNA plasmid is 72% G+C rich (as opposed to 30 to 40% G+C rich for the others). This may explain why the consensus origin sequence was not found, although a sequence bearing structural but not strict sequence similarity to the pC194 origin is present upstream of the Rep open reading frame (bp 1327 to 1269 of the published pIJlOl sequence [39]). It is not known whether this sequence has origin activity.


M-Os.
The M-Os of several ssDNA plasmids have been analyzed (4a, 12, 14, 24). The minimal sequences in Call cases are large (at least 130 to 2'0 bp) and contain imper-fect palindromic structures. All M-Os show orientation-dependent activity. DNA sequences of three unrelated M-Os are presented in Fig. 5, in which pCalindromes are indicated.
orientation 방향

Conversion is mediated by host-encoded factors (4a, 24). Replication is initiated in each of the three types of M-Os in vivo by host-encoded ribonucleic acid (RNA) polymerase. since rifampin blocks conversion (4a; L. Boe and A. Gruss, unpublished data).
RNA polymerase is known to initiate replication at the M-Os of the filamentous ssDNA phages (2). The properties of an ssDNA plasmid in different hosts may depend on whether the M-O is active.
Many of the M-Os are host specific (4a. 12, 24); only the M-O of pUBIIO is known to function in more than one host (4a). In all hosts, a plasmid lacking an active M-O is still viable, but aIccumulaltes ssDNA; conversion in these cases initiates only nonspecifically, at a reduced frequency (4a, 24, 77). In some hosts, such as S. alacl)is (24), Streptococcus I(eul.olnliOtielC (12), and Streptomvces litidlans (13), deletion of the M-O also causes decreased plasmid copy number and, in the first two cases, pronounced plasmid segregational instability. In contrast, the M-O does not affect plasmid copy numbei in Bacillits suubtilis (4a, 14, 24).