PLASMID REPLICATION
Copy Number Control and Its Role in Segregational Stability
Maintaining an upper limit to plasmid copy number may be necessary to ensure the viability of the host. In addition, to prevent plasmid loss, copy number controls must also respond rapidly in a cell inheriting few copies. This is pa1rticularlty important in the absence of a separate partitioning locus (par).
No evidence exists for a par locus on the ssDNA plasmids, in the context of its original definition, i.e., membrane attachment sites that physically aid segregation of sufficient numbei-s of plasmids such thcat they become estab lished in each dLughter cell (37). In plasmid pTX81, the only muta-tions that seem to affect plasmid stability are those that caffect replication tunctions (19, 20, 24. 35: A. Gi-uss and R. Novick, unpnLblished data) (see below). Ihe par function, which is repo-ted to reside on plasmid pLS11 (10) and which is also founLd in placsmids pTA1060 (5) and pBAA1 (14), was later revealed (on pBAA1) to be the M-O (14).
In ssDNA plasmids, stability seems to be coupled with replication and not with acldiscrete par-like function. Known copy number control elements affect the initiation step. Other modes of copy number control may act at other steps. e.g.. elongation or conversion of ssDNA to dsDNA.
The rate of elongation can be strongly affected by the DNA sequence and consequently can affect the plasmid copy number; this was demonstrated by observing the effects of insertion of a 22-bp termination sequence into pUC plasmids (27). Similarly, the efficiency of initiation of conversion of ssDNA to dsDNA can determine the plasmid copy number (Fig. 2). It remains to be shown that these steps can be manipulated for copy control. Most of the data presented below on copy control at the level of initiation derive from plasmid pT181. The informa tion is presented here with the speculation that analogous systems of copy control and stability are operational on other ssDNA plasmids.
Regulation of Rep synthesis.
Plasmid copy number control by modulation of Rep expression has been extensively studied for pT181 (see reference 67 for a review). Regulation is achieved by the production of countertranscript (CT) RNA, which modulates either transcription or translation of the rep messenger RNA (mRNA), or both, and keeps the copy number within a limited range.
Certain point mutations affecting the CT RNA have increased copy numbers (9, 64). Mutants in which the CT RNA is not synthesized have a much increased copy number (50-fold [9, 641), but are not lethal. This suggests that other factors prevent runaway replication. Some of these are as follows: (i) certain high copy-number plasmids are accompanied by deletion deriva tives, which may bind to and thus deplete Rep (34); (ii) host factors (possibly rate limiting) besides Rep regulate the plasmid copy number and might prevent runaway replication (34); (iii) if ssDNA is accumulated at high concentrations, it could interfere with replication by titrating Rep (Rep binds to ssDNA [44]); (iv) Rep, as shown for E. coli plasmid R6K (79), may be less active at high concentrations; (v) the origin may be refractive to overreplication (see below). When a daughter cell inherits too few copies of a plasmid, the imbalance is rapidly adjusted by overreplication (28, 63).
This is expected if the CT RNA (which is present in large quantities [67] and would therefore be equipartitioned) de cays rapidly. In this case, it would not inhibit plasmid replication in daughter cells (see below). Overreplication of plasmids in cells receiving too few copies is a sensitive control that responds to rapid fluctuations in copy number.
Availability of origin as a mechanism of copy number control.
Several factors affect the efficiency with which an origin is recognized. Increased transcription through the pBR322 origin in the direction opposite to that of replication (81) reduces replication efficiency, whereas transcription through oriC enhances replication (3).
Plasmid superhelicity affects the efficiency of replication initiation in pBR322 (55). Furthermore, hemimethylated pBR322 (the product of a round of replication) is not recognized as a substrate for replication (78). Therefore, if the necessary effectors are present but the origin is not readily available, initiation may be inefficient. Such factors may have a large effect on plasmid copy number and hence on copy control. A novel locus, comp, affecting the efficiency of origin utilization, has recently been identified on plasmid pT181 (19, 20, 35).
A comp mutant plasmid in S. aureus is unable to compete successfully for Rep in the presence of a comp+ plasmid and requires a longer period than the wild-type plasmid to repopulate a cell if its copy number is reduced and then derepressed (20). comp is thought to facilitate Rep recognition of the origin, possibly by affecting the super-he licity of origin sequences (19). An analogous IOCuS WaIS also identified in pUB110 (72; J. Polak, unpublished data). Nor mally, pT181 (co/p is located about 1,200 bp from the origin: its activity is orientation independent and is inversely pro portional to its distance from the origin (20).
How can copy number control affect plasmid segregational stability?
A simple proposal by Novick and co-workers presents an alternative to an independent partitioning system (20, 28, 63): plasmids, if partitioned randomly, should be present at random copy numbers in daughter cells after cell division. If a plasmid has a tight copy control system (e.g., high turnover rate of CT RNA), cells receiving fewer plas mid copies will rapidly adjust this situation by synthesizing Rep protein and undergoing compensatory overreplication (28, 63).
However, if Rep protein is synthesized too slowly (e.g., owing to transcriptional repression of repC mRNA by an overactive CT RNA) or if the origin is not readily available for replication (e.g., owing to a (comp defect), this adjustment may not occur fast enough to replete the plasmid copy pool. The copy number will remain low in these cells. and plasmids may be lost in subsequent cell divisions. Since the activity of comp is inversely proportional to its distance from the origin (20). it follows that cloning of foreign DNA segments into an ssDNA plasmid diminishes the interaction between comp and the origin and hence lowers the ability of the plasmid to fadjust its copy number upon cell division (20). Failure to rapidly correct copy number fluctuation may account for the reported segr-ega tional instability of these plasmids when they are used as cloning vectors (6).
INFIDELITY OF RCR
The replication of ssDNA plasmids does not always follow the normal system schematized in Fig. 2. Nearly every step in the process is either known or proposed to digress from its usual function, thus effecting rearrangements.
In addition. RCR generates ssDNA; in every recombination process, ssDNA is a reactive intermediate.
The rearrangements and the possible steps at which they occur are described below. Numbering follows the replication steps schemaitized in Fig.2
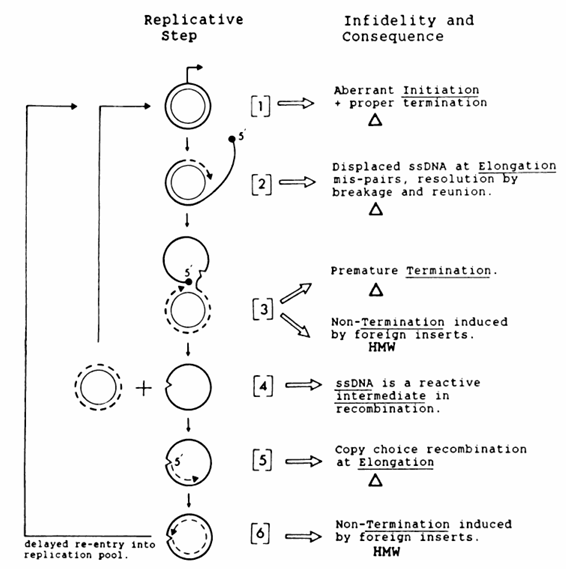
Step 1: initiation. Initiation at a pseudo-plus origin, fol lowed by termination at the correct plus origin, provides a consistent explanation for the endpoints found in al deletion induced by pC194 replication (53).
pseudo 가짜의, 허위의
Step 2: elongation and displacement during plus-strand synthesis. Elongation during plus-strand synthesis with con comitant displacement of plus-strand ssDNA may lead to initiation of homologous recombination by the displaced strand (52). Deletion frequencies between long direct repeats carried by ssDNA plasmids are about 1,000-fold greater than deletion frequencies of equivalent constructions in the chro mosome (61). Insertion of an ssDNA plasmid in the B. smibtilis chromosome also stimulates deletion and amplifica tion of adjacent repeat sequences (62: M.-A. Petit. J. Mesas Mesas, P. Noirot, and S. Ehrlich, submitted for publication)
Step 3: termination of plus-strand synthesis. Terminaition of plus-strand synthesis by a wobbly recognition of the termi nation sequence results in the formation of recombintants. Either the wrong sequence is recognized, which pr-ovokes precocious termination (step 3A). or the correct termination sequence is ignored (step 3B). In step 3A, precocious termination of replication in ssDNA plasmids, recombinants are generated. In certain E. coli plasmid construCts comprising the ssDNA filamentous phage. pC194. and pBR322 sequences, the nick site at the phagge replication origin is a deletion hot spot in E. coli (54). Likewise, in certain of these constructs, a deletion hot spot also exists within the pC194 sequence (53).
It was hypothe sized that this hot spot was the replication nick site (53), and it was later confirmed diirectly that this hot spot was in the pC194 origin region within an 18-bp sequence containing the nick (22). The other deletion endpoints were at originlike sequences (53. 54). T'he simple explanation for the formation of these deletions is the correct initiation aIt the origin and aberrant termination at sequences resembling the termina tion-origin sequence (22: M.-F. Gros, H. te Riele, and S. D. Ehrlich. submitted for publication). 'I'he 18-bp sequence containing the pC194 nick site is also present in pUBIIO (22). A cointegrate between pC194 and pUBIIO replicons was previously reported to decombine at high frequencies at precisely this sequence (26). Studies by Gros et al. showed that the cointegrate is actually resolved by initiation at one origin and accurate and efficient termi nation Cat the 18-bp sequence of the second origin (22) (i.e., initiation at the pC194 origin and termination in the pUBIIO 18-bp sequence. and vice versa). Although 55 bp is required for Rep to recognize the pC194 origin, only 18 bp is required for Rep to mediate normal termination (22). Likewise, al though pUBIIO requires a larger origin for initiation, only 18 bp is necessary for normal termination (4a).The termination reaction can be degenerate, and so to study the termination process, a plasmid was constructed comprising the entire pC194 origin sequence rand an insertion of the 18-bp termi nation sequence. In this plasmid, termination occurred pre maturely at the 18-bp sequence. It was shown that certain base changes within the 18-bp sequence still provoked termination, albeit at reduced frequencies; thus, errors by the Rep protein in termination reSulted in deletions (Gros et al.. Submitted). In step 3B, the correct termination sequence is ignored. The presence of certain insertions of foreign DNA in any position within the ssDNA plasmids results in the formation of high-molecular-weight tandem multimers (HMW) (23). HMW are produced in the wild-type host with any ssDNA plasmid vectors, but not with a vector apparently outside this family (plasmid pAM31 was tested). It is hypothesized that the presence of a foreign DNA insertion interferes with normal termination of plus-strand replication. Notably, all shuttle vectors tested consisting of an ssDNA plasmid and pBR322 sequences produce HMW in gram-positive hosts; for these constructs, more than 70)c> of the plasmid DNA is present as HMW.
Step 4: release of circular ssDNA. Released circular ssDNA may be a reactive recombination intermediate, with possible physiological effects such as SOS induction (H. te Riele, S. D. Ehrlich. and R. D'Ari, unpublished data). Results of preliminary experiments suggest that ssDNA stimulates in termolecular homologous recombination (V. Vagner, per sonal communication).
Step 5: elongation during minus-strand synthesis. Elonga tion during minuS-Stralnd synthesis is subject to slipped mispairing recombination. Recombination, tested by using short (9-bp) direct repeats flanking an inactive transposon (i.e., inverted repeat sequences), is stimulated 150- to 1,500 fold in plcasmids that replicate via an ssDNA intermediate compared with the chromosome or plasmids which do not gener-ate ssDNA (38: Jatnni&re and Ehrlich. submitted). A Study of different-length direct repeats flanking inverted repealts in ssDNA plasmids in B. sotbtili.s showed that deletion frequencies are proportional to the length of the repeats (69). A copy choice recombination mechanism has been proposed (see reference 17 for a discussion), and results of a model system developed with E. coli give good evidence of this mechanism (7). A difference should be noted, however, between B. slub tilis and E. c(oli recombinations that are or are not stimulated by ssDNA. In E. (oli, deletions between short repeats can occur at about the same frequencies as deletions between long repeats (D. Brunier, personal communication). In B. sulbtilis, deletion frequencies of short direct repeats are 105 to 106 times lower than deletion frequencies of long direct repeats (38). This implies that the copy choice mechanism of recombination, which is active in the deletion of short direct repeats (7), may be less frequent or better monitored in B. slubtilis than the equivalent mechanism in E. coli.
Step 6: termination of minus-strand synthesis. The forma tion of HMW (step 3B) was also proposed to occur by a failure to properly terminate minus-strand synthesis. If the 3'-OH end is displaced, a nonterminating RCR would con tinue to form HMW.